【NGS with Data Science】Gene2vec distributed representation of genes pipeline reproduction

This article explains how to use the pipeline in the paper "Gene2vec: distributed representation of genes based on co-expression." and re-train the model with your data.
Install pipeline
First, we create the path to download the package . We created a conda virtual environment of Python 3.7 as following commands:
mkdir gene2vec_test
cd gene2vec_test/
conda create -n gene2vec_test_env python=3.7
conda activate gene2vec_test_envThen download the package with git command:
git clone https://github.com/jingcheng-du/Gene2vec.git
cd Gene2vec/Because there are differences in parameter names between gensim 3.x and 4, if you want to use gemsim with 3.x version, remember to change requirements.txt: change gensim>=3.4.0 to gensim==3.4.0. (If you want to use gensim with 4.x version you need to modify gene2vec.py, such as changing the "size" of the word2vec object to "vector_size", etc.)
pip install -r requirements.txtAfter the installation program is completed, we can test it as the command:
cd src/
python gene2vec.py
If the following message appears, the installation should be good.
usage: gene2vec.py [-h] N [N ...]Training Models
Then we could use the testing data to run the example :
python gene2vec.py ../data/ ../out txtThe gene vector would saved at the running path and here were some example data:
head -n 2 outgene2vec_dim_100_iter_1.txt
FGF6 0.002153057 0.001094015 0.0040485994 0.003507802 -0.0034948308 -0.0032273065 0.002758114 -0.0044576144 0.002355916 0.0017780543 0.004745546 0.0018376901 -0.0035088449 -0.0005739574 -0.000108827386 0.002103943 -0.0038852852 0.0012951874 -0.0031769034 -0.004375249 0.004074314 -0.0026881285 0.004214152 -0.004282877 0.0022233215 0.004169825 0.00061325595 2.0139367e-05 -0.0016913096 0.0025811284 0.0031880501 -0.0019990925 -0.0047910786 0.002188197 0.0026727102 -0.0006805879 0.00019095051 0.0010278132 0.0017754859 0.0031797176 -0.003708027 -0.0043337652 0.0035265626 -0.0008643125 0.00084504695 -0.00054039893 0.0003750502 -0.0037928058 -0.0042927195 -0.0047074244 -0.0017722481 0.00025958134 -0.0026379086 0.00018871028 -0.0019723917 -0.00021585514 0.0033635853 0.0022829815 -0.0024485104 0.0011425553 0.003241704 0.0047381823 0.0012685822 0.0041412427 0.0019761408 0.0019880526 0.0039201365 0.0013327249 -0.002263571 -0.0044547706 0.0037608626 0.00095062394 -0.00030630908 0.0031630904 -0.0018972668 0.004344254 0.0025073248 0.0037321039 -0.004189576 0.0025266777 0.0005846647 0.0019490473 0.0018105969 -0.004199487 0.0020253006 -0.0017606984 -0.004815944 0.0046018823 0.0042982115 0.00051282457 -0.0009345786 0.003392324 -0.0032844574 0.0011845101 -0.0011895953 -0.0012602699 -0.00042309787 0.004582391 0.0025786795 -0.0024350516
GFI1B -0.0029350498 0.0043180487 -0.004318311 0.0019120751 -0.0038370104 -0.00023128637 -0.004420749 -0.0035758333 -0.0040116534 0.0012707855 -0.0009630754 0.0004477923 0.0020208724 -0.00041648198 0.003939566 -0.0040858993 -0.004756729 0.0018472039 -0.0021072265 0.002428173 -0.00014559152 0.0045682737 -0.0033070655 -0.0035072211 0.00053472363 -0.0026147643 0.00052187295 0.0034156216 -0.0035089792 0.001963524 -0.0040159533 0.0029510746 0.004897053 0.0017880275 0.0009832341 -0.004501591 -0.0021778357 0.002407189 0.000616764 -0.003227798 -0.0042902012 0.0024847183 0.003374102 0.002082069 0.001478934 0.0048288074 0.0042617135 -0.0018422379 0.0039390987 0.00026498176 -0.00028904268 0.0011463418 0.0027650178 0.0037835115 0.0007013022 0.004905474 0.0006962089 0.0002940799 0.0038201583 -0.0031658853 -0.00292867 -0.00054527074 0.004884007 0.002188833 0.00015647558 -0.002252723 0.0020673836 0.0038181976 0.00041569016 -0.003276892 -0.002797324 0.0020927635 0.0010414731 -0.004298761 0.002510277 -0.0017390802 0.00439754 -0.0042876415 -0.00071369467 0.002830168 -0.0037963414 0.0036242604 0.00023945107 0.004529737 0.001412234 -0.0010020512 0.0044706156 0.0015063612 0.0029264004 -0.0043842485 -0.0016326424 0.0022118944 0.00042738195 -0.004558031 -0.003733534 -0.0029223813 0.0048098615 -0.0019367941 0.00491898 -0.0025868856 In addition, we could use the pre-trained model and run the tSNE, for example:
pip install MulticoreTSNE
pip install scikit-learn
python tsne_multi_core.pyand use plot.py to plot it :
import pandas as pd
import matplotlib.pyplot as plt
plt.style.use('ggplot')
df=pd.read_csv("TSNE_data_gene2vec.txt_100.txt",sep=" ",header=None)
df.columns = ['x', 'y']
plt.scatter(x=df["x"],y=df["y"])
plt.show()than could get the 2d projection of the genes.
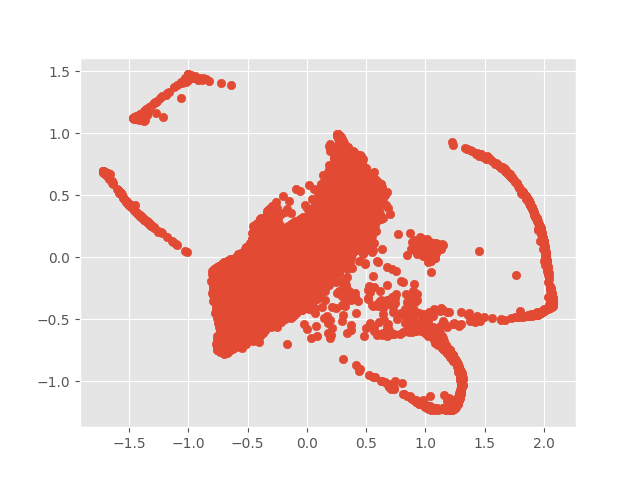
Each data point was the representation of a gene. Now, we have roughly completed the conversion of gene names to vectors for decentralized representation. This method can enhance the prediction capabilities of other biological markers .
If we trace the source code of gene2vec.py, can see that the training method is similar to the general NLP word vector method. The idea was only to change the general NLP input to the gene list of the GSEA data set.
References
- Du, J., Jia, P., Dai, Y. et al. Gene2vec: distributed representation of genes based on co-expression. BMC Genomics 20 (Suppl 1), 82 (2019). https://doi.org/10.1186/s12864-018-5370-x
A passionate bioinformatician focuses on the next generation of medical science and biotechnology.
喜歡這樣的教學創作的話,歡迎 小額贊助 給予支持🙏


