【3D Genome】Computer vision-based methods to predict Hi-C Topologically associating domains (TAD) - A proof of concept.

A Hi-C contact map can be represented as a 2d array (matrix) and visualized as a gray-scale image. Topologically associating domains (TAD) were identified as a high interaction region inside a submatrix. Therefore, we can transfer the Hi-C contact map into image space and then use the common computer vision technique to filter the TAD just like how we find ROI.
Methods
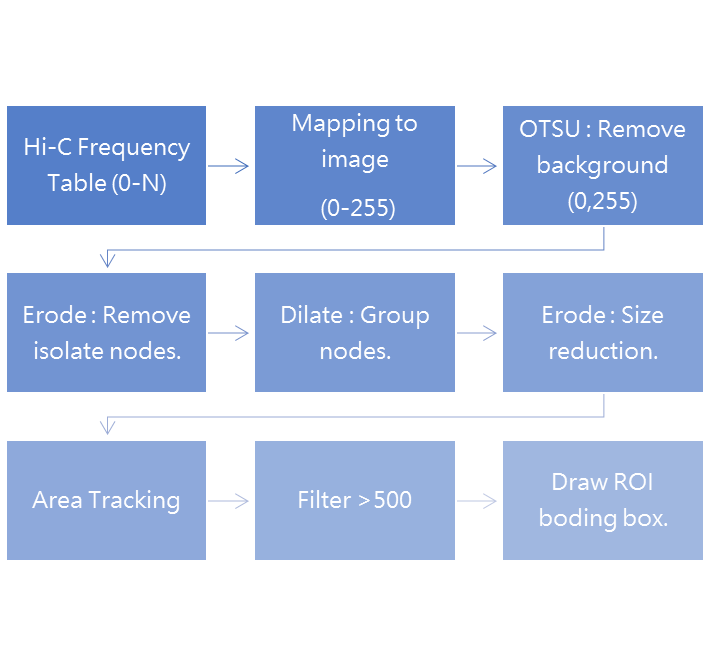
The analysis pipeline is illustrated in the figure below, which is based on traditional computer vision methods to detect ROIs.
Experiment & Discussion
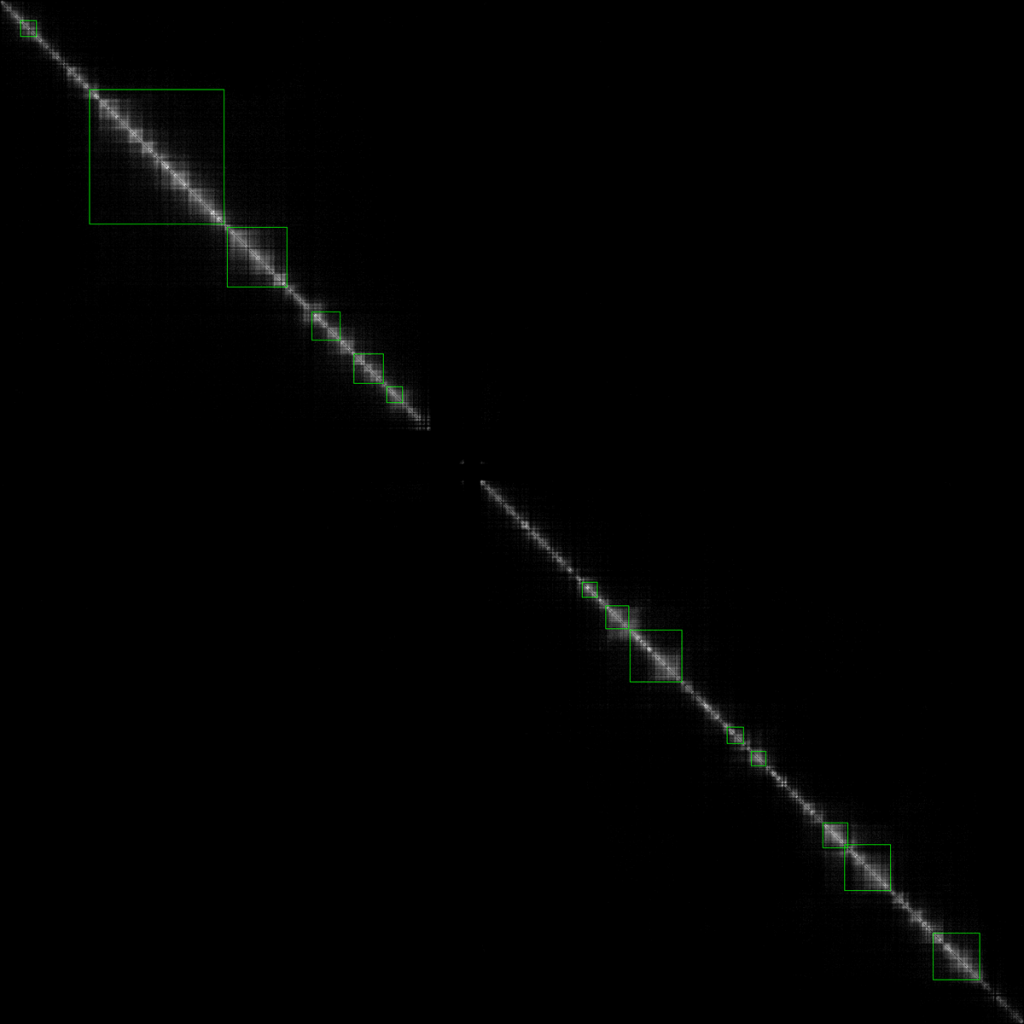
The result is as follows:

and the final TAD boundary was marked by the green line.
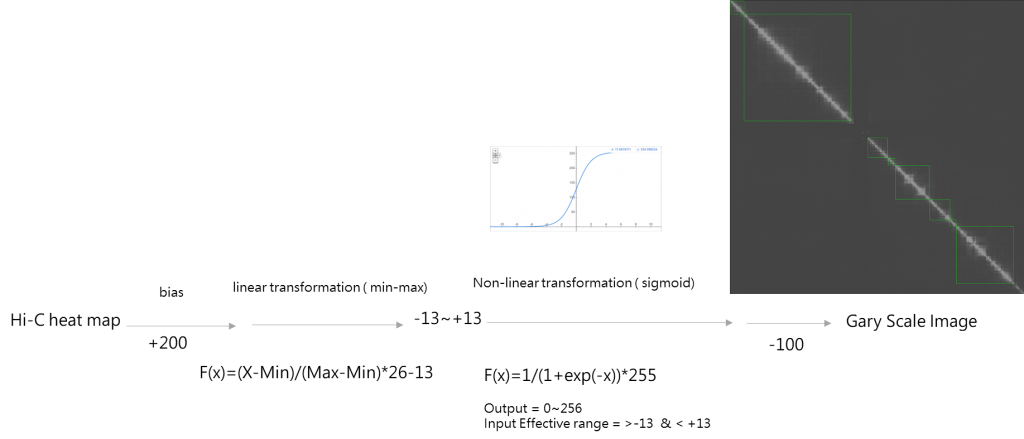
After some trial and error, we know that the transfer of the Hi-C contact map into an image should have some parameters, and an equation similar to the perceptron is introduced.
Code Implementation
HiCCVTAD.java
package com.whuang022.bio;
import org.bytedeco.javacpp.opencv_core.Mat;
import org.bytedeco.javacpp.opencv_core.MatVector;
/**
* HiC TAD DTO Class
* @author whuang022
*/
public class HiCCVTAD{
public Mat TADImage;
public MatVector TADRects;
}HiCCV.java
package com.whuang022.bio;
import org.bytedeco.javacpp.BytePointer;
import org.bytedeco.javacpp.opencv_core;
import static org.bytedeco.javacpp.opencv_core.IPL_DEPTH_8U;
import org.bytedeco.javacpp.opencv_core.IplImage;
import org.bytedeco.javacpp.opencv_core.Mat;
import org.bytedeco.javacpp.opencv_core.MatVector;
import org.bytedeco.javacpp.opencv_core.Scalar;
import static org.bytedeco.javacpp.opencv_core.cvCreateImage;
import static org.bytedeco.javacpp.opencv_core.cvGetSize;
import static org.bytedeco.javacpp.opencv_imgproc.COLOR_GRAY2BGR;
import static org.bytedeco.javacpp.opencv_imgproc.CV_CHAIN_APPROX_NONE;
import static org.bytedeco.javacpp.opencv_imgproc.CV_RETR_EXTERNAL;
import static org.bytedeco.javacpp.opencv_imgproc.CV_THRESH_BINARY;
import static org.bytedeco.javacpp.opencv_imgproc.CV_THRESH_OTSU;
import static org.bytedeco.javacpp.opencv_imgproc.boundingRect;
import static org.bytedeco.javacpp.opencv_imgproc.cvDilate;
import static org.bytedeco.javacpp.opencv_imgproc.cvErode;
import static org.bytedeco.javacpp.opencv_imgproc.cvtColor;
import static org.bytedeco.javacpp.opencv_imgproc.findContours;
import static org.bytedeco.javacpp.opencv_imgproc.rectangle;
import static org.bytedeco.javacpp.opencv_imgproc.threshold;
/**
* HiC JavaCV Process Class
* @author whuang022
*/
public class HiCCV {
public static Mat HiCtoMat (HiC hic,int shiftVal){
IplImage image = IplImage.create(hic.getRowSize(),hic.getColSize(), IPL_DEPTH_8U, 1);
Mat mImage=new Mat(image);
for(int i=1;i<hic.getRowSize();i++){
for(int j=1;j<hic.getColSize();j++){
BytePointer bytePointer = mImage.ptr(i,j,0);
double val=hic.get(i,j);
double color= ((val - hic.getMin()) * (1/( hic.getMax() - hic.getMin()) * 255)); //linear mapping HiC to Gay Scale Image (0-255)
bytePointer.putInt((int)color+shiftVal);
}
}
return mImage;
}
public static HiCCVTAD HiCTADCallingMorphology (HiC hic,int shiftVal,int min){
Mat HiCMat;
Mat otsu = new Mat();
IplImage otsuIp ;
IplImage colseIp ;
Mat HicMatOut = new Mat();
HiCMat=HiCCV.HiCtoMat(hic,shiftVal);
MatVector contours = new MatVector();
MatVector outcontours = new MatVector();
threshold(HiCMat, otsu, 10, 255, CV_THRESH_OTSU + CV_THRESH_BINARY); // OTSU to Mark Acctivate Zone
otsuIp = new IplImage(otsu);
colseIp= cvCreateImage(cvGetSize(otsuIp), IPL_DEPTH_8U, 1);
//Opening Operation
cvErode(otsuIp, colseIp, null, 1);
cvDilate(colseIp, colseIp, null, 3);
cvDilate(colseIp, colseIp, null, 1);
Mat colse=new Mat(colseIp);
findContours(colse, contours,CV_RETR_EXTERNAL, CV_CHAIN_APPROX_NONE);
cvtColor(HiCMat,HicMatOut, COLOR_GRAY2BGR);
for(int i=0;i<contours.size();i++){
opencv_core.Rect mr = boundingRect(contours.get(i));
if(mr.area()>min){
rectangle(HicMatOut, mr, new Scalar(0, 255, 0, 1));
outcontours.put(mr);
}
}
HiCCVTAD out =new HiCCVTAD();
out.TADImage=HicMatOut;
out.TADRects=outcontours;
return out;
}
}
HiC.java
package com.whuang022.bio;
import java.io.BufferedReader;
import java.io.FileNotFoundException;
import java.io.FileReader;
import java.io.IOException;
import java.util.ArrayList;
/**
* HiC Data Reader Class
* @author whuang022
*/
public class HiC {
private ArrayList<ArrayList<Double>> data = new ArrayList<ArrayList<Double>>();
private double maxVal;
private double minVal;
public int getRowSize(){
return data.size();
}
public int getColSize(){
return data.get(0).size();
}
public double get(int row,int col){
return data.get(row).get(col);
}
public double getMax(){
return maxVal;
}
public double getMin(){
return minVal;
}
public HiC(String filePath) throws FileNotFoundException, IOException{
BufferedReader reader = new BufferedReader(new FileReader(filePath));
maxVal=Double.MIN_VALUE;
minVal=Double.MAX_VALUE;
String line ;
while((line=reader.readLine())!=null){
String[] rowData = line.split(" ");
ArrayList<Double> row=new ArrayList<>();
for (String rowData1 : rowData) {
double val=Double.parseDouble(rowData1);
row.add(val);
if(val>maxVal){
maxVal=val;
}
if(val<minVal){
minVal=val;
}
}
data.add(row);
}
}
}
Conclusion
The final result is as follows which shows the approximate TAD boundary.
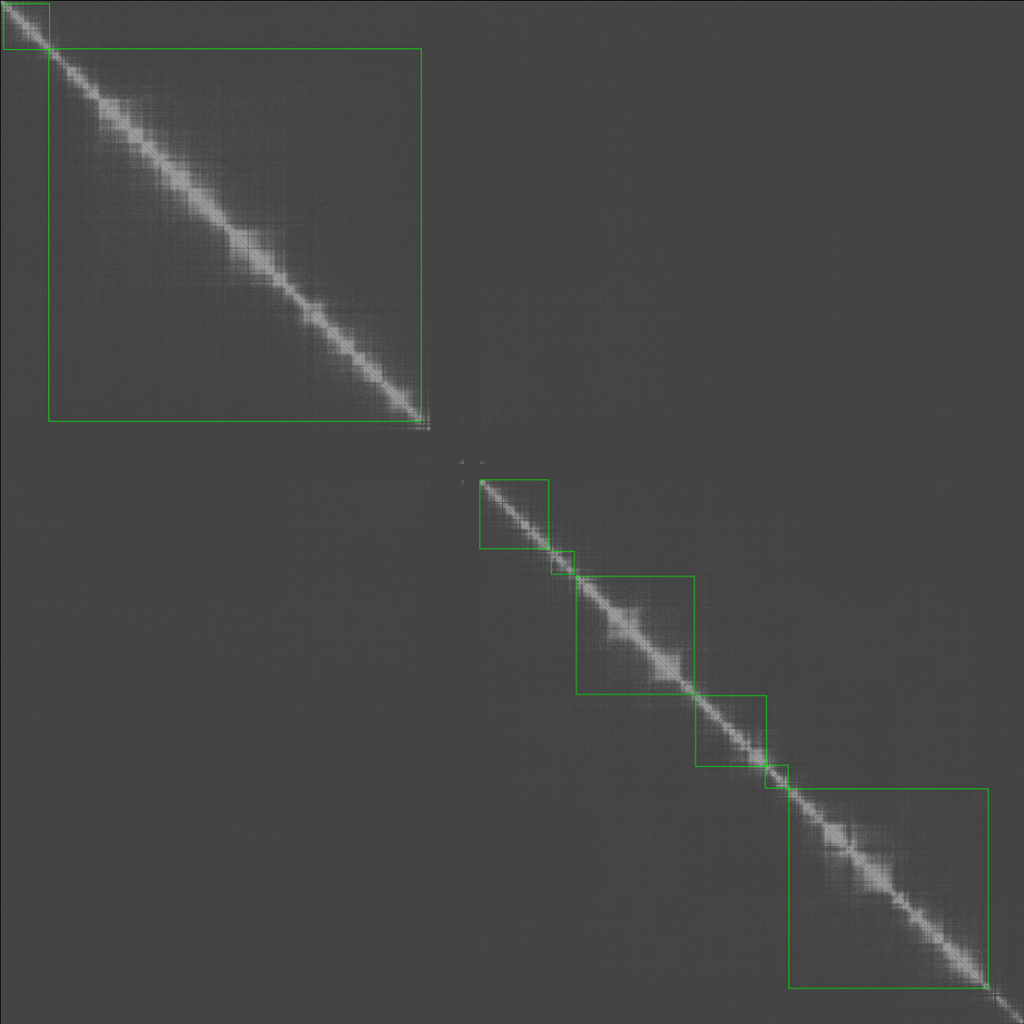

The above is a TAD calling algorithm proof of concept. In the future, we can compare it with the main TAD calling methods.
A passionate bioinformatician focuses on the next generation of medical science and biotechnology.


